July 8th
Finally got in with IT to talk about VPN issues that have preventing WFH. Looks like a security update happened for the VPN that makes it no longer compatible with my computer as my 2016 macbook can’t update to the neccessary MacOS 13 Ventura. I suppose that WFH will now have to be reserved for another day (and another paycheck…)
July 9th
Went into the office and worked on some rphil updates. Reduced results to gene level by e value, and worked to filter DESeq results. Updated figures and enrichment.
July 10th
Met with mac to go over rphil updates. Next steps are to continue to mess around with different filter settings to see what genes persist, and keep updating visualizations.
July 17th
Worked on DESeq and Heatmap code to 1) clean it up to make it a bit more clear and 2) refine the heatmaps for each of the analysis so far. Started first readthrough of the rphil manuscript too.
July 19th
Light week, but got some technical issues figured out and moved enough of the clam repo local to do some of the less heavy analysis from home again. Finished manuscript read through. Prepped for Chris’ panel next week.
July 24th
Went back and reconciled for the missing 193 sample. Redid featurcounts to include 193.
July 25th
Contunued reconcilation for 193 and went and redid the analysis downstream of hisat / featurecounts. The number of DEGs now more closely matches Giles’ results. First round of prefiltering of deseq results too.
July 26th
Chris’ student panel thing. More work on deseq prefiltering
July 27th
More deseq prefiltering and moved onto by group specific prefiltering. Identified consistent top genes across each pre filtering parameter and set up list for annoation.
July 30th
Post filtering analysis, creating heatmaps, and running recurring DEGs through annotation.
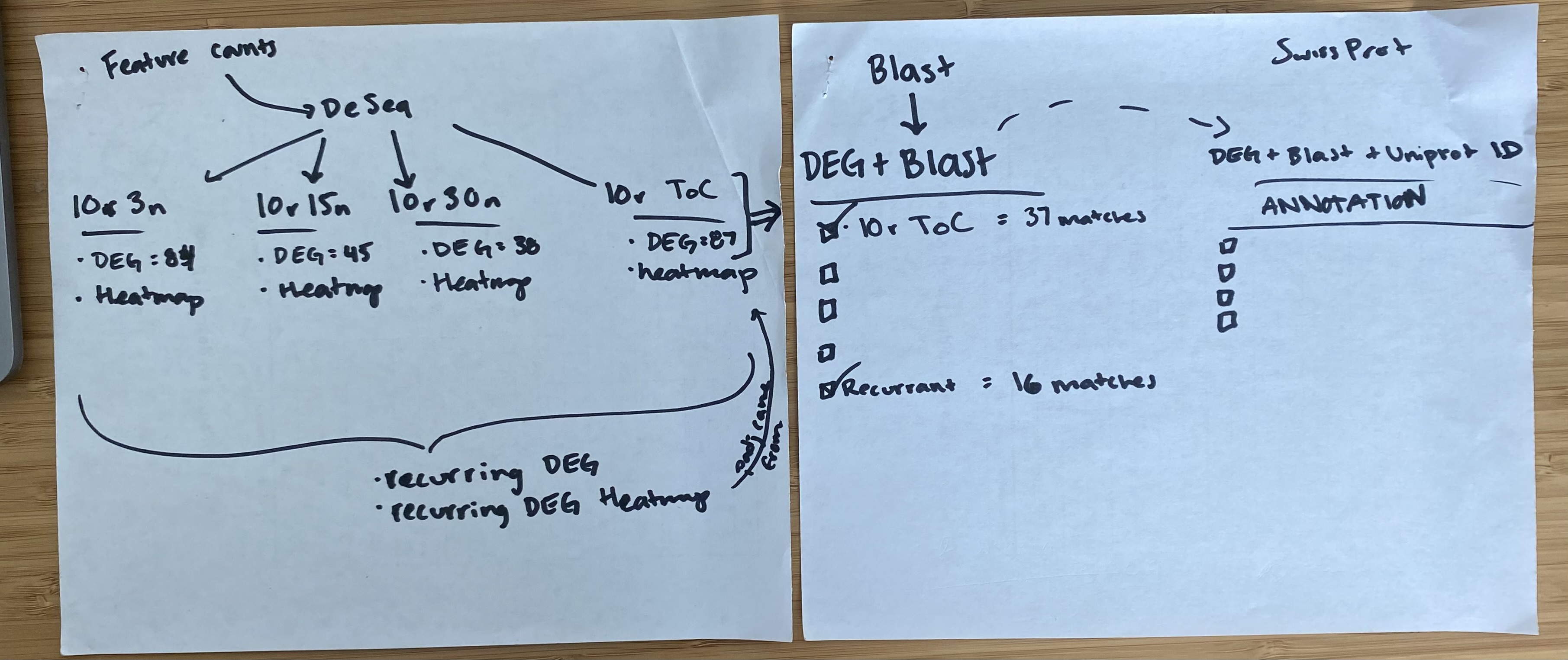
^ basically a short visual summary of the filtering process
pre-filtered featurecounts matrix / deseq to only include results that hit certain parameters:
average of 10+ reads across 3+ samples (10r3n) (DEG = 84)
average of 10+ reads across 15+ samples (10r15n) (DEG = 45)
average of 10+ reads across 30 samples (10r30n) (DEG = 38)
average of 10+ reads across each treatment (10r ToC) (DEG = 87)
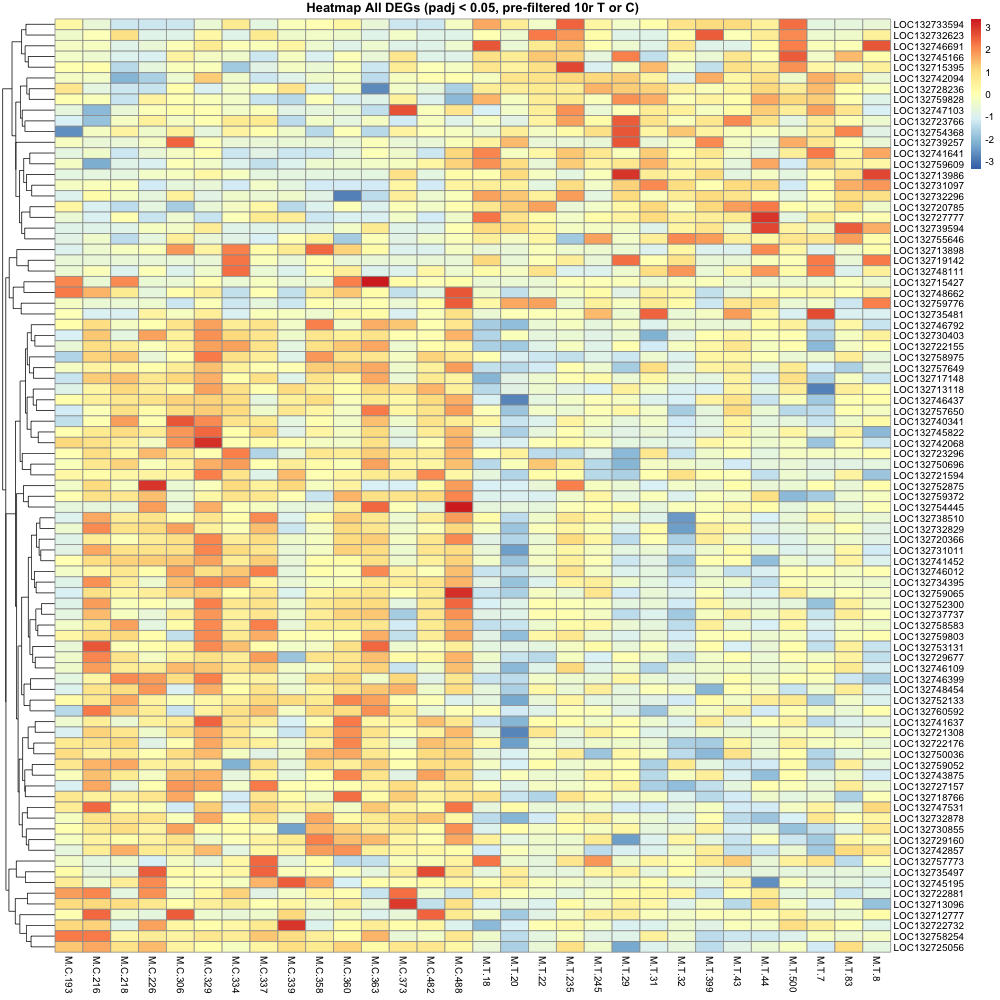
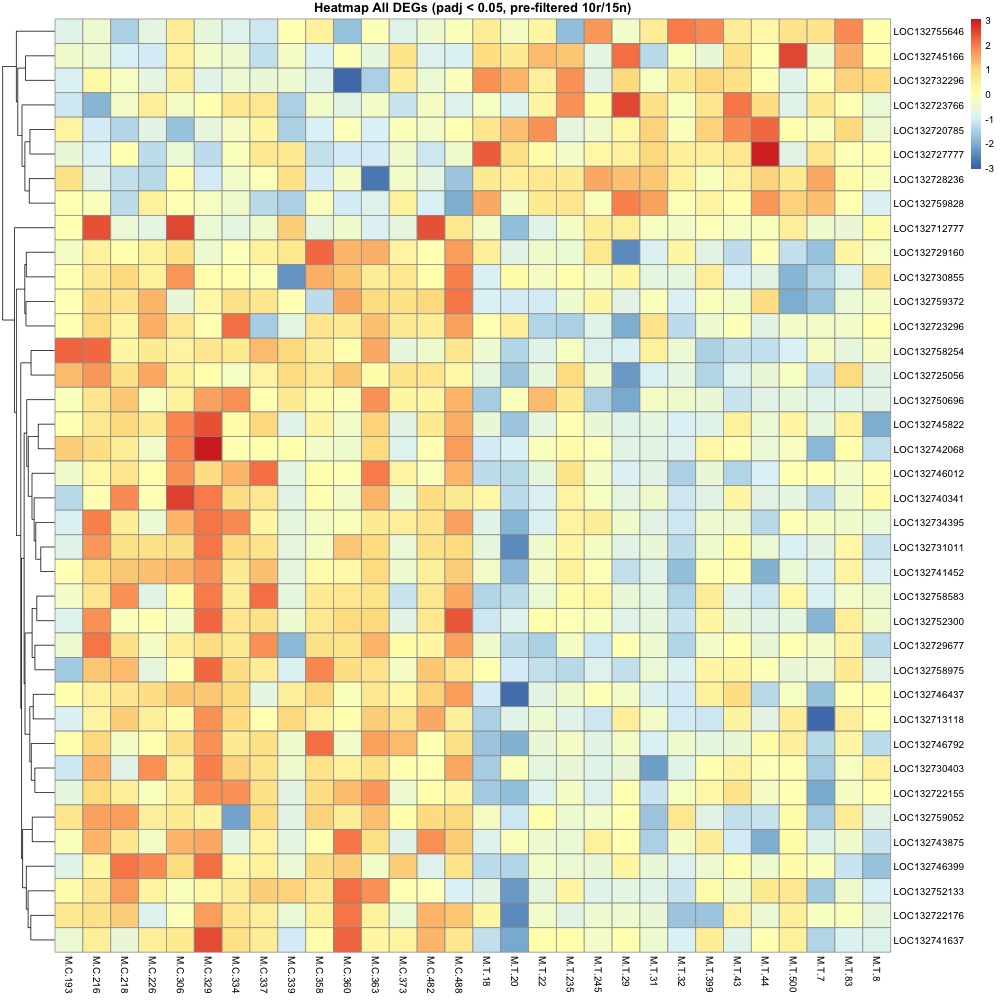
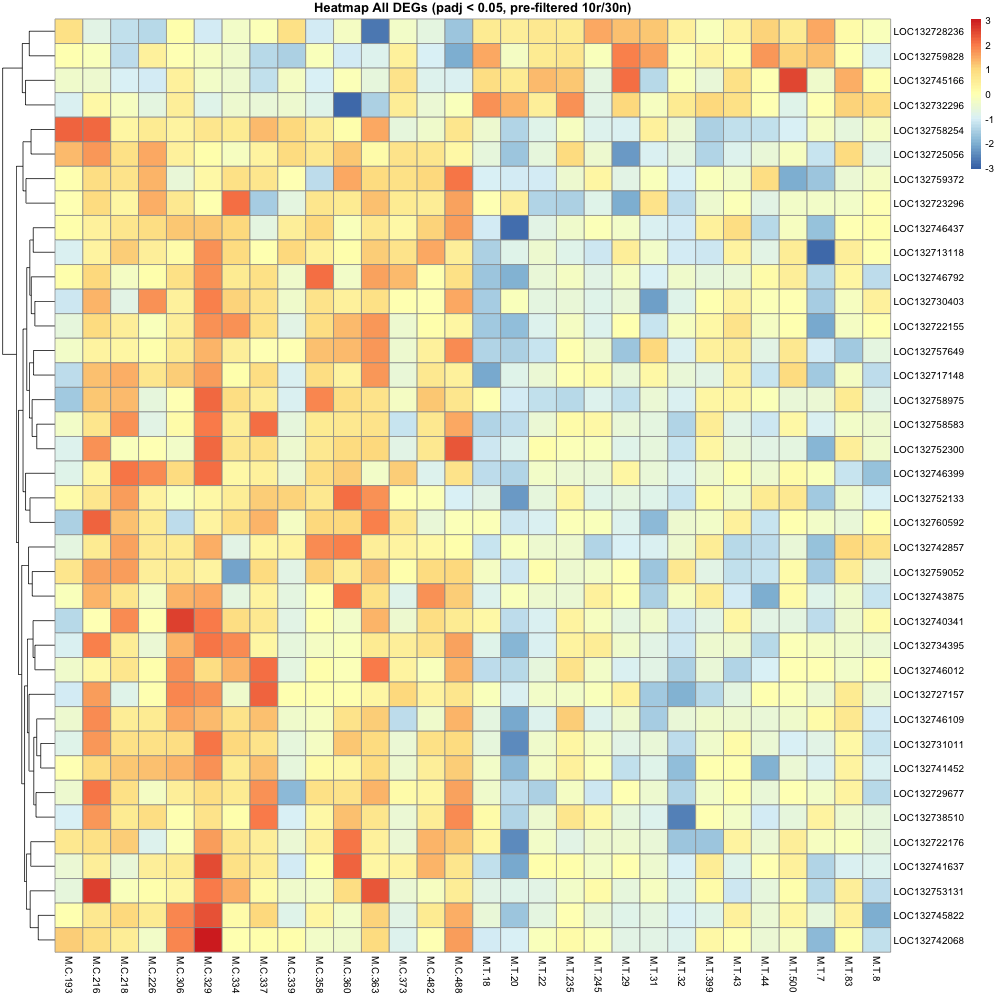
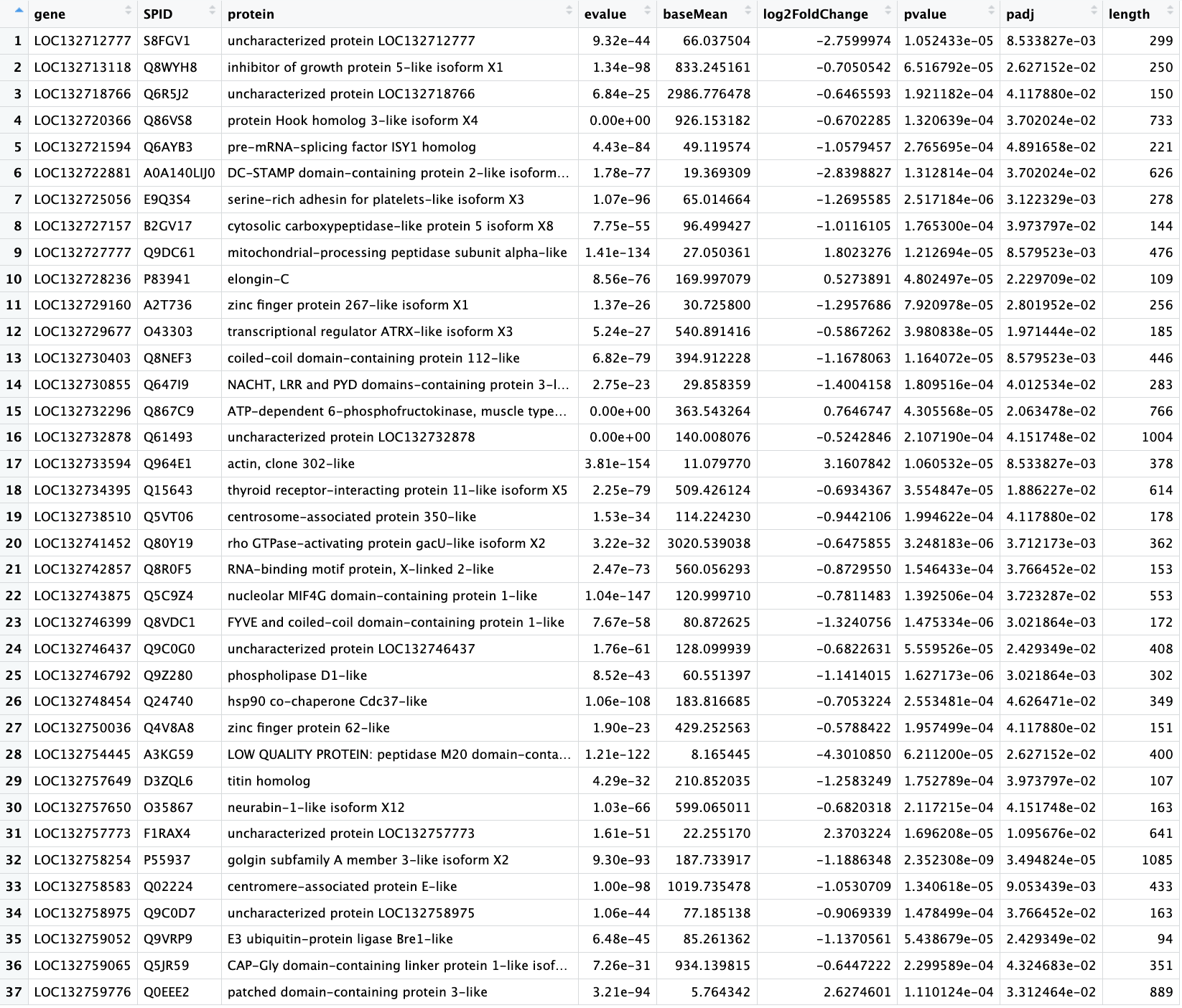
After making heatmaps, I used the list of recurring genes I made and ran that though the first annotation step. (Not going all the way through uniprot just yet )
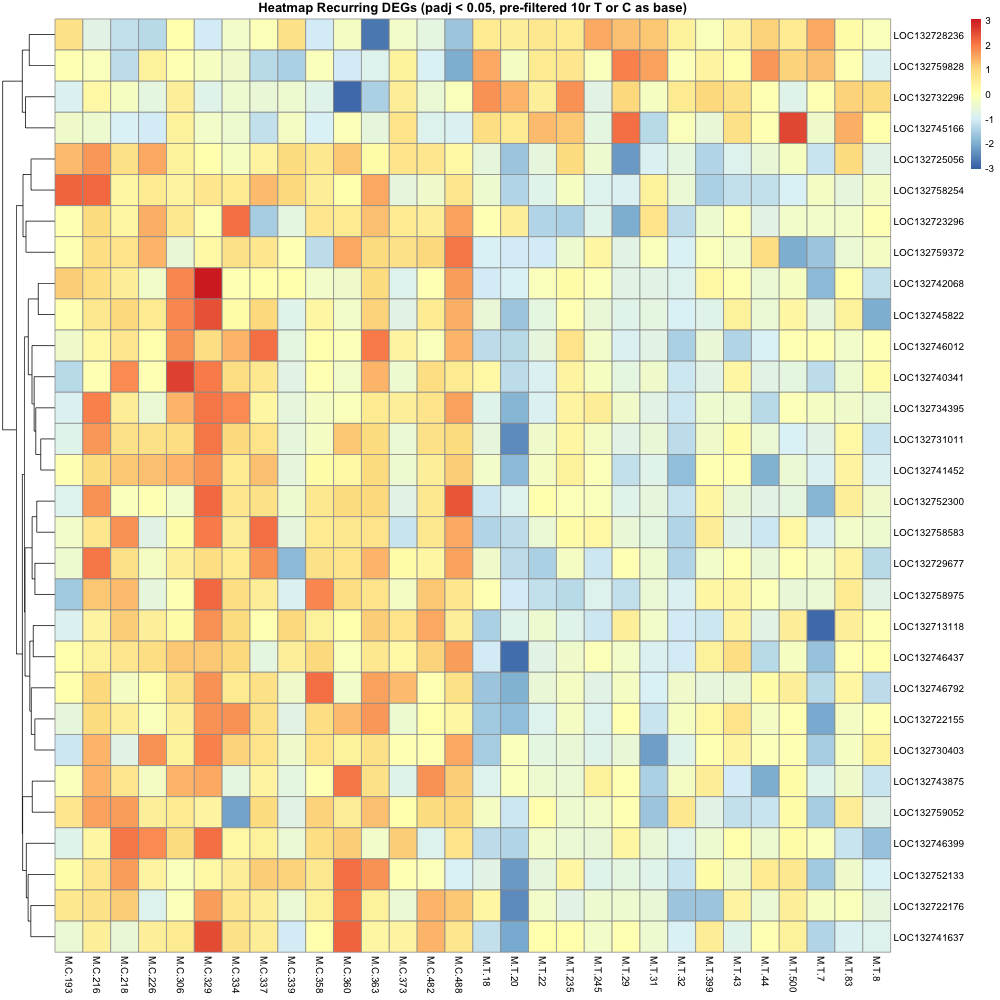
Then made a heatmap of these recurring genes
July 31st
Met with mac to review results and discuss new parameter options for pre filtering. Then learned how to read gonad histology slides with Mac and Eala